VolSurf+
The pharmacokinetic behaviour of compounds is linked to their efficacy and thus is critical for drug discovery. Understanding how to optimise compounds according to multiple simultaneous criteria is a great advantage in focusing design efforts.
VolSurf+ creates 128 molecular descriptors from 3D Molecular Interaction Fields (MIFs) produced by our software GRID, which are particularly relevant to ADME prediction and are also simple to interpret. One example would be the interaction energy moment descriptor between hydrophobic and hydrophilic regions, which is important for membrane permeability prediction. These can then be used with provided chemometric tools to build statistical models.
VolSurf+ also comes with a number of models that we have developed using both public and pharmaceutical data, including passive intestinal absorption, blood-brain barrier permeation, solubility, protein binding, volume of distribution, and metabolic stability.
A more complete description of the original VolSurf method and derived models is available in the references section.
Key Features:
- Calculate ADME relevant descriptors and perform statistical modelling using experimental data
- Predict the behaviour of new compounds based on new or established ADME models
- Interactively sketch structural modifications to optimise ideas according to multiple criteria
- Select compounds with similar ADME properties to a query structure
Package content
VolSurf+ Modeller
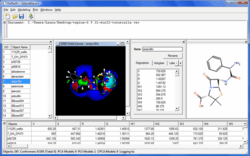
- Conformational search and internal hydrogen bond search for increased accuracy
- 128 ADME relevant descriptors
- Visualise GRID fields to aid descriptor interpretation
- Up to 18 graphical analysis methods to aid model interpretation
VolSurf+ Designer
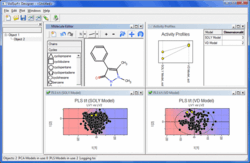
- Access existing custom or standard models
- Project new compounds simultaneously into multiple models
- Sketch compound changes for interactive optimization according to multiple criteria
VolSurf+ Selector
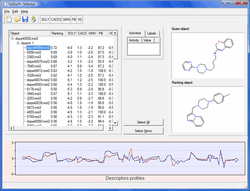
- Perform virtual screening to find new compounds with similar ADME properties to a query structure
Availability
VolSurf+ is available for Windows® and Linux® operating systems.
Editions comparison
VolSurf+ comes in two editions: Solo and Suite
VolSurf+ Solo provides interactive interfaces for ADME modeling, design, and virtual screening; VolSurf+ Suite provides unlimited interface access and also command-line utilities for wider enterprise deployment in cheminformatics processes.
References
-
Sara Tortorella, Emanuele Carosati, Giulia Sorbi, Giovanni Bocci, Simon Cross, Gabriele Cruciani, Loriano Storchi
Combining machine learning and quantum mechanics yields more chemically aware molecular descriptors for medicinal chemistry applications
J. Comp. Chem., 2021 - Molecular Fields in Quantitative Structure-Permeation Relationships: the VolSurf Approach
J. Mol. Struct.: THEOCHEM, 2000, 503, pp. 17-30 - VolSurf: a new tool for the pharmacokinetic optimization of lead compounds
Eur. J. Pharm. Sci., 2000, 11, pp. S29–S39 - Predicting Blood-Brain Barrier Permeation from Three-Dimensional Molecular Structure
J. Med. Chem., 2000, 43 (11), pp. 2204–2216 - Use of MIF-based VolSurf descriptors in Physicochemical and Pharmacokinetic Studies
in Molecular Interaction Fields, 2005, Wiley, pp. 173-196 - Molecular Interaction Fields in ADME and Safety
in Molecular Interaction Fields, 2005, Wiley, pp. 45-82 - Prediction of Oral Drug Permeability
in Rational Approaches to Drug Design, 2001, Prous Science, Barcelona, pp. 271–280
